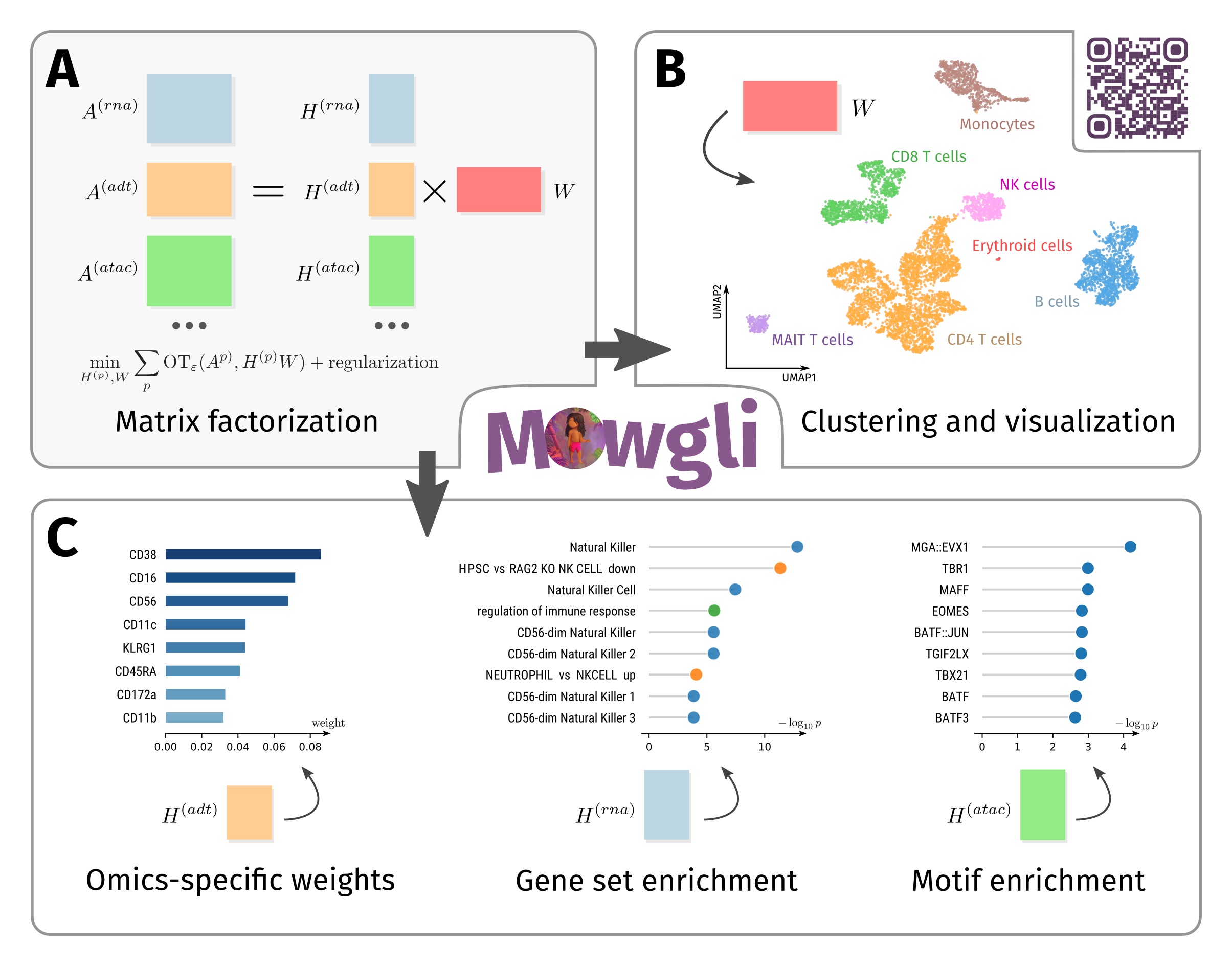
Mowgli: Multi Omics Wasserstein inteGrative anaLysIs#
Mowgli is a novel method for the integration of paired multi-omics data with any type and number of omics, combining integrative Nonnegative Matrix Factorization and Optimal Transport. Read the preprint here and fork the code here!
Install the package#
Mowgli is implemented as a Python package seamlessly integrated within the scverse ecosystem, in particular Muon and Scanpy.
via PyPI (recommended)#
pip install mowgli
via GitHub (development version)#
git clone git@github.com:cantinilab/Mowgli.git
pip install ./Mowgli/
Getting started#
Mowgli takes as an input a Muon object and populates its obsm and uns fiels with the embeddings and dictionaries, respectively. Visit the Getting started and API sections for more documentation and tutorials.
You may download a 10X Multiome demo dataset at https://figshare.com/s/4c8e72cbb188d8e1cce8.
from mowgli import models
import muon as mu
import scanpy as sc
# Load data into a Muon object.
mdata = mu.load_h5mu("my_data.h5mu")
# Initialize and train the model.
model = models.MowgliModel(latent_dim=15)
model.train(mdata)
# Visualize the embedding with UMAP.
sc.pp.neighbors(mdata, use_rep="W_OT")
sc.tl.umap(mdata)
sc.pl.umap(mdata)
Citation#
@article{huizing2023paired,
title={Paired single-cell multi-omics data integration with Mowgli},
author={Huizing, Geert-Jan and Deutschmann, Ina Maria and Peyre, Gabriel and Cantini, Laura},
journal={bioRxiv},
pages={2023--02},
year={2023},
publisher={Cold Spring Harbor Laboratory}
}